CSCS Benchmarks Molecular Dynamics Codes on GPGPU Systems
Earlier this year, CSCS hosted a tutorial called “Molecular Dynamics Codes on GPGPUs” — it provided an overview of classical molecular dynamics software, showed some benchmarking results, and gave a hands-on walkthrough using GPU-accelerated MD codes.
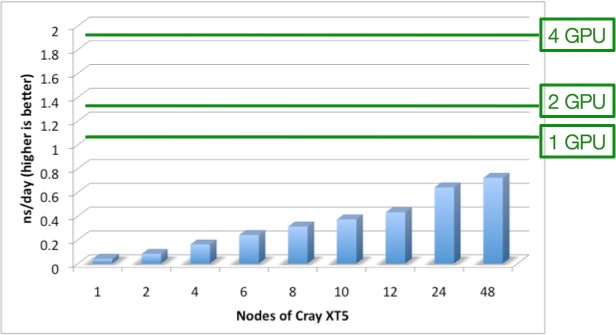
CSCS published a brief summary of the benchmarking results that were featured in the tutorial. CSCS looked at two of the most popular MD codes at CSCS: NAMD (v2.7) and AMBER (v11) – both already have CUDA implementations.The goal of the benchmark is to check if the GPU-based system’s time-to-solution would be decreased, and to see if the implementations were mature enough for production science.